This page lists published computer applications, including console, web and window-based applications.
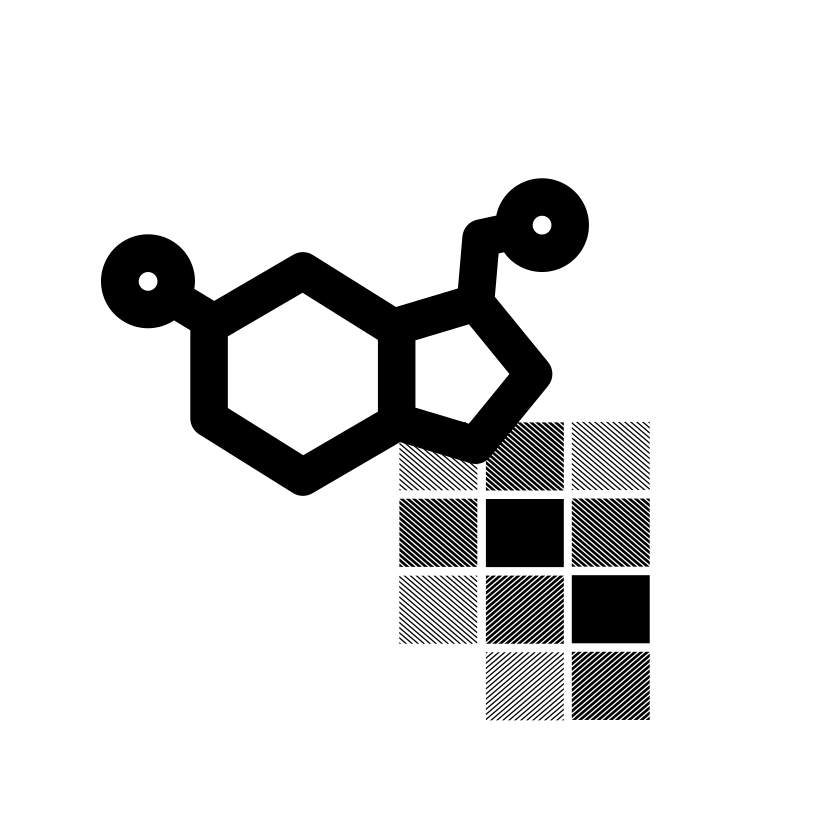 |
iCAN is a console application that enables molecular encoding of any organic compound using the Interpretable Carbon-based Array of Neighborhoods method. It offers interpretable representations tailored for machine learning, outperforms previous methods in peptide and protein classification, and aids in drug discovery and disease diagnosis. [Publication 1, Publication 2]. |
 |
PolarDiagrams is an interactive console application that enables the evaluation of complex models across various domains, including biology, medicine, climatology, and machine learning. It addresses the challenge of evaluating models using multiple metrics by providing an integrated visualization approach. [Publication]. |
 |
DNAsmart is an interactive and visual analytical application that enables the evaluation and ranking of DNA storage encoding approaches based on multiple attributes. It provides an accessible and tractable way to solve comparison, ranking, and validation tasks related to DNA storage encoding. [Publication]. |
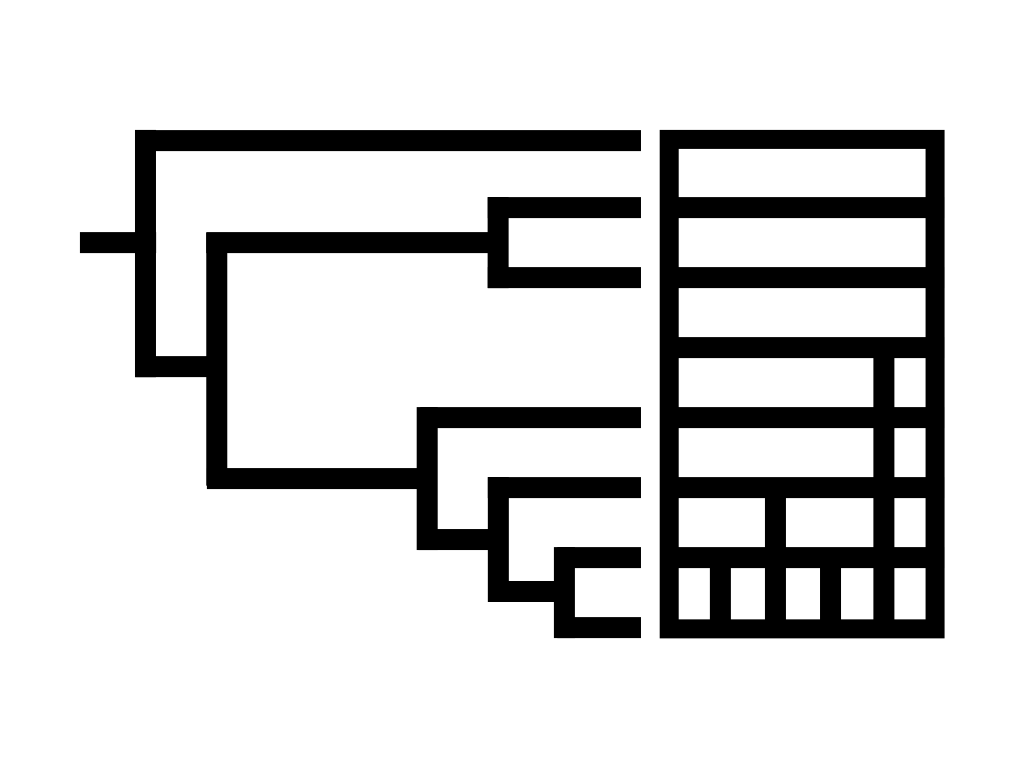 |
CAPT is an interactive web application that supports users in exploration and validation tasks related to phylogeny-based taxonomy. It bridges the gap between phylogeny and taxonomy by offering two simultaneous views: a phylogenetic tree view and a taxonomic icicle view. [Publication]. |
 |
MOVIS is a multimodal omics application that enables the import, embedding, clustering, and interactive visualization of different types of omics data with temporal information. It provides insight into patterns and anomalies in an integrated environment, complementing traditional data exploration approaches. [Publication]. |
 |
PeptideREACToR is a web and console application for peptide encoding performance in the biomedical domain. It provides a baseline of 48 encoding groups tested on 50 datasets, enabling initial encoding selection, objective comparison of new encodings, and sophisticated encoding optimization in automated machine learning pipelines. [Publication]. |
 |
MOSGA is a versatile web application that combines genome annotation and comparative genomics capabilities. It provides a user-friendly interface to generate and integrate annotations, analyze results in a genome browser, and enable comparative genomics analyses on high-quality genome assemblies. [Publication 1, Publication 2]. |
 |
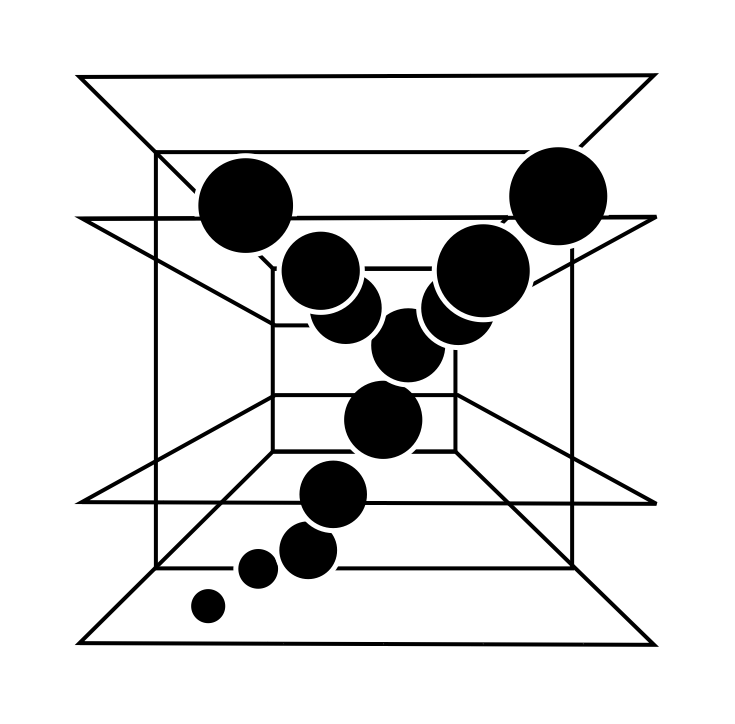
modelAR is a modular and evaluative framework to improve Augmented Reality (AR) visualization for surgical applications. It enables testing different visualization techniques and presets for displaying 3D biomedical data in an AR environment. [Publication]. |
 |
SeeVIS is a console application that enables automated and qualitative visualization of time-lapse microscopy data. It offers a solution to the lack of visualization alternatives for rapid qualitative characterization of microbial colonies using a space-time cube. [Publication]. |









